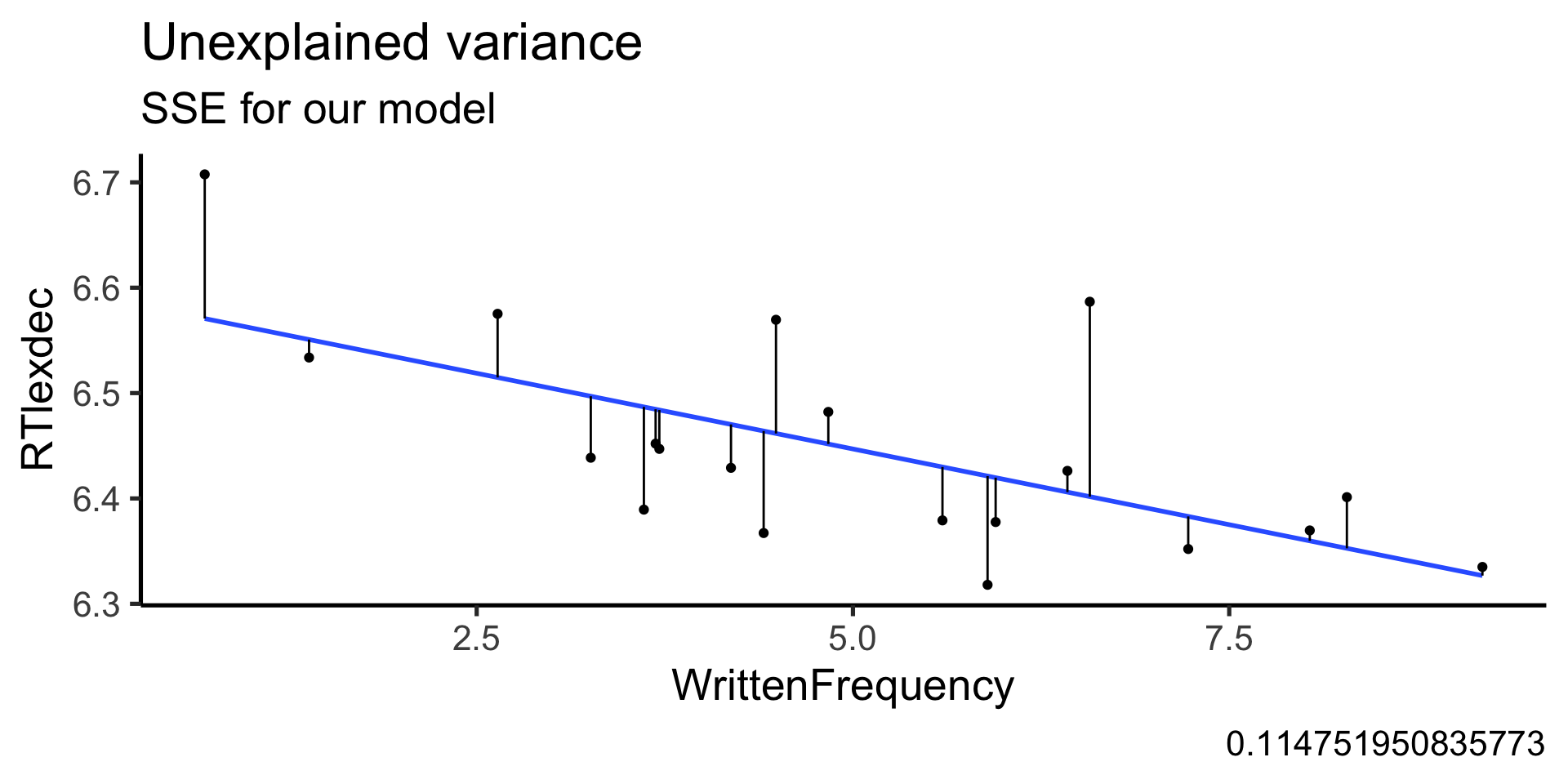
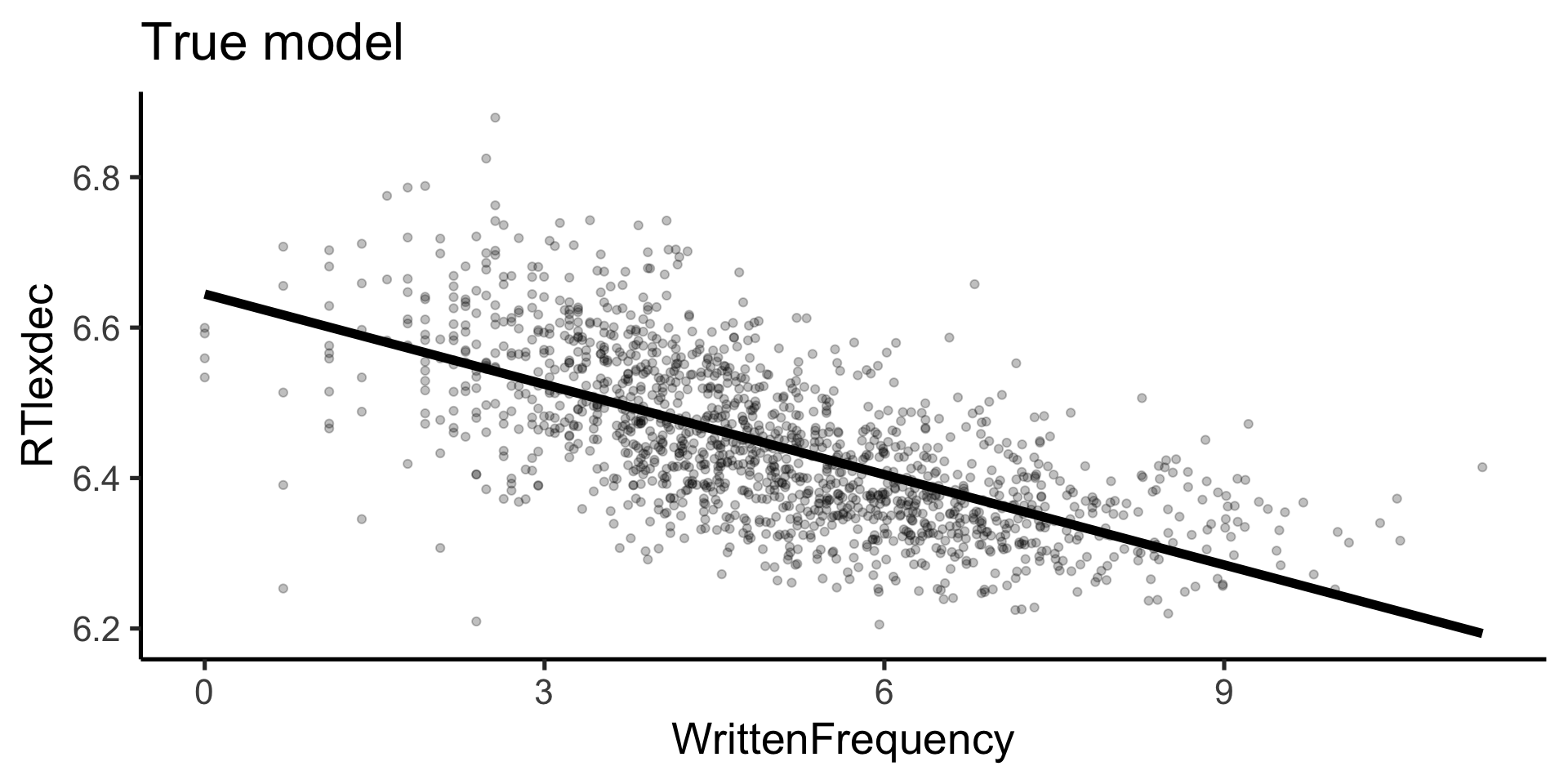
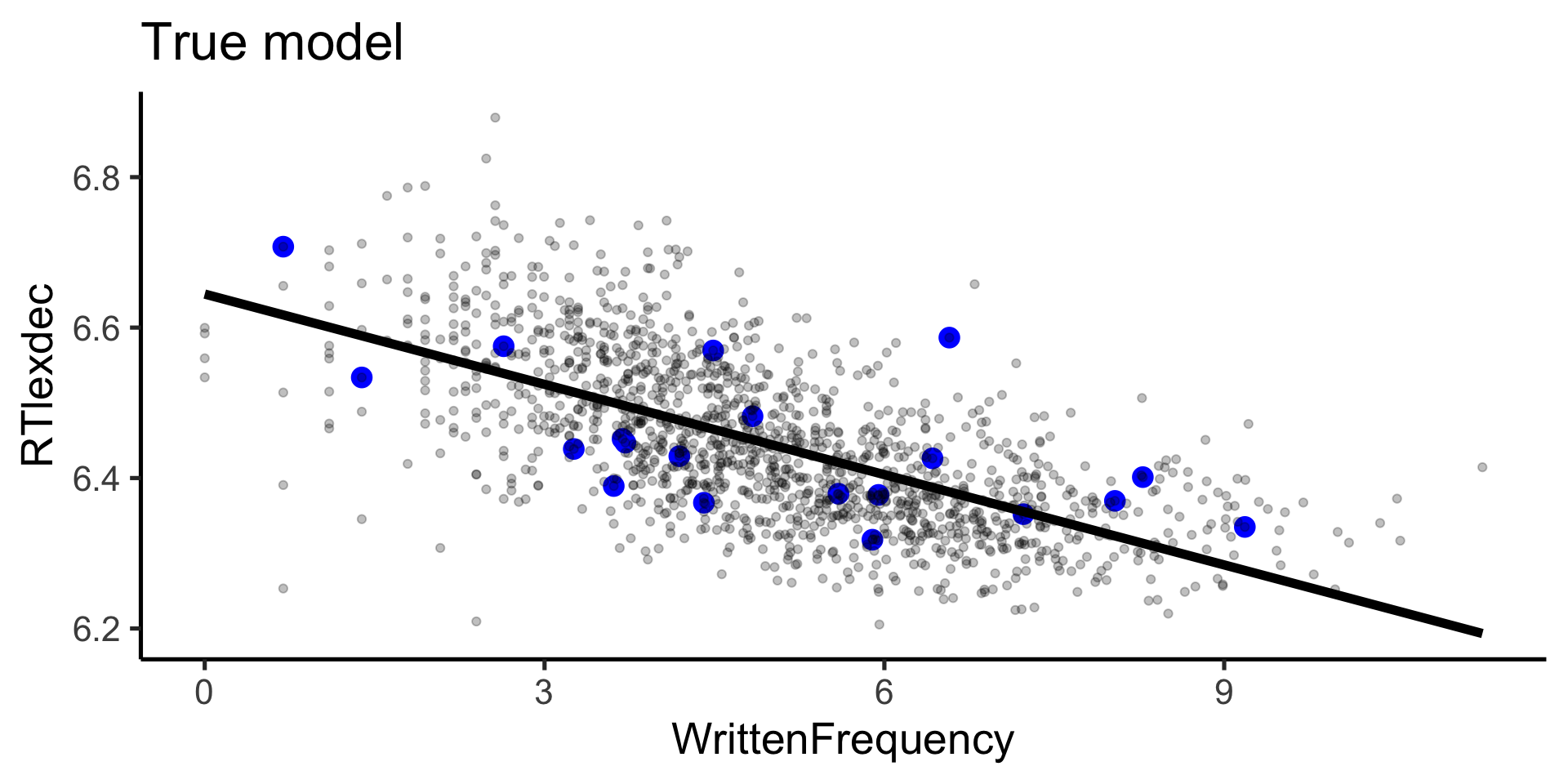
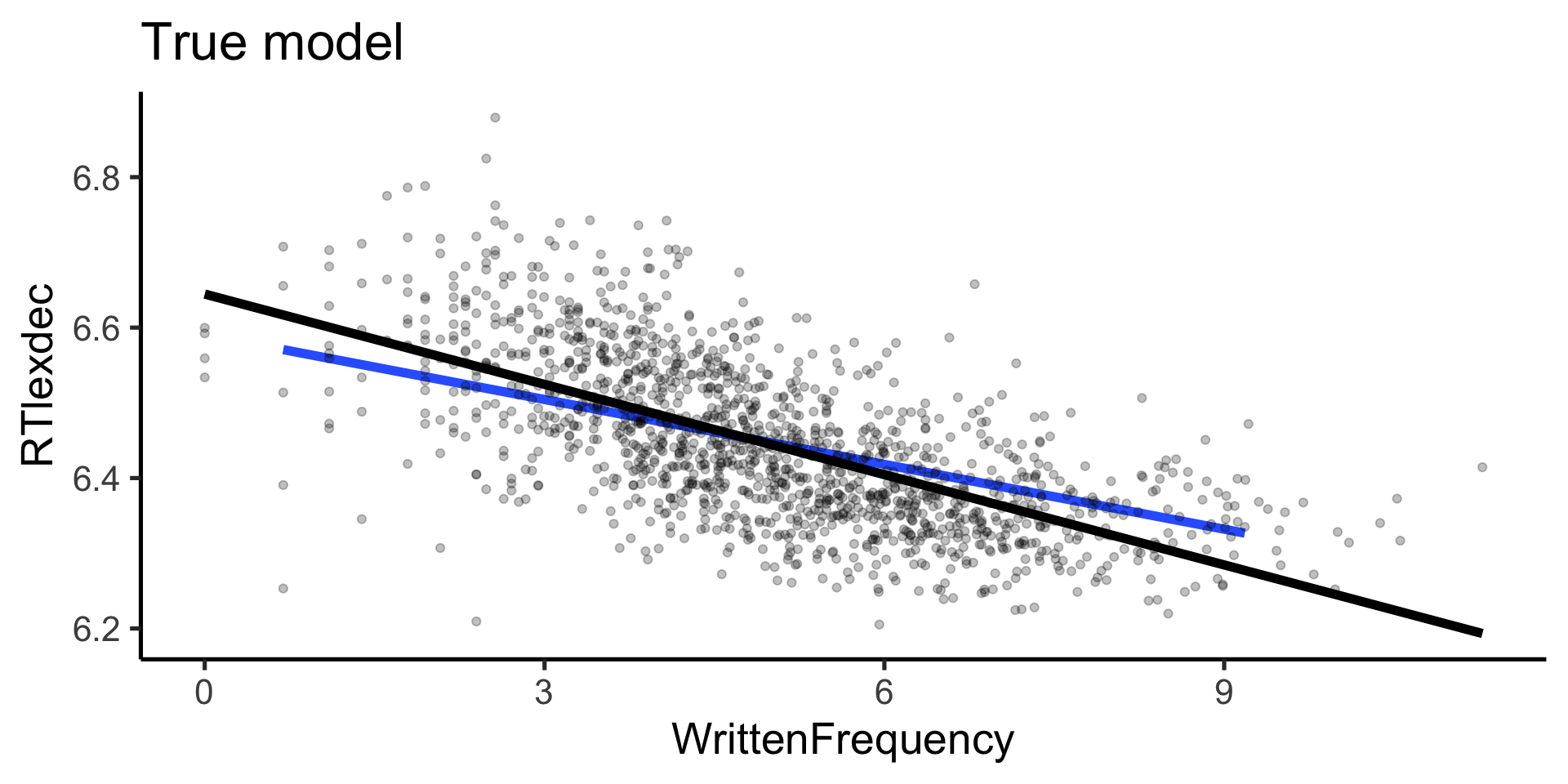
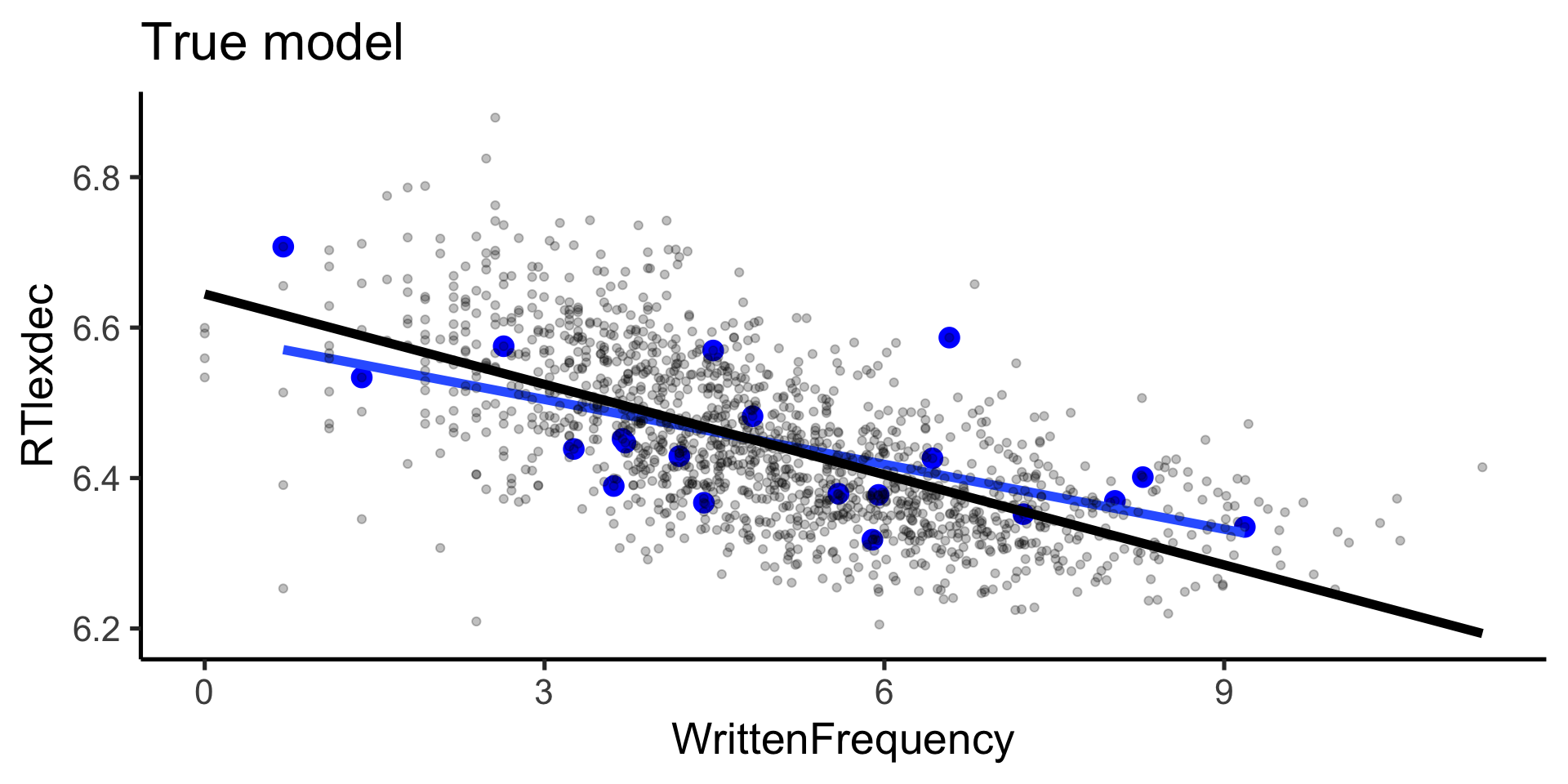
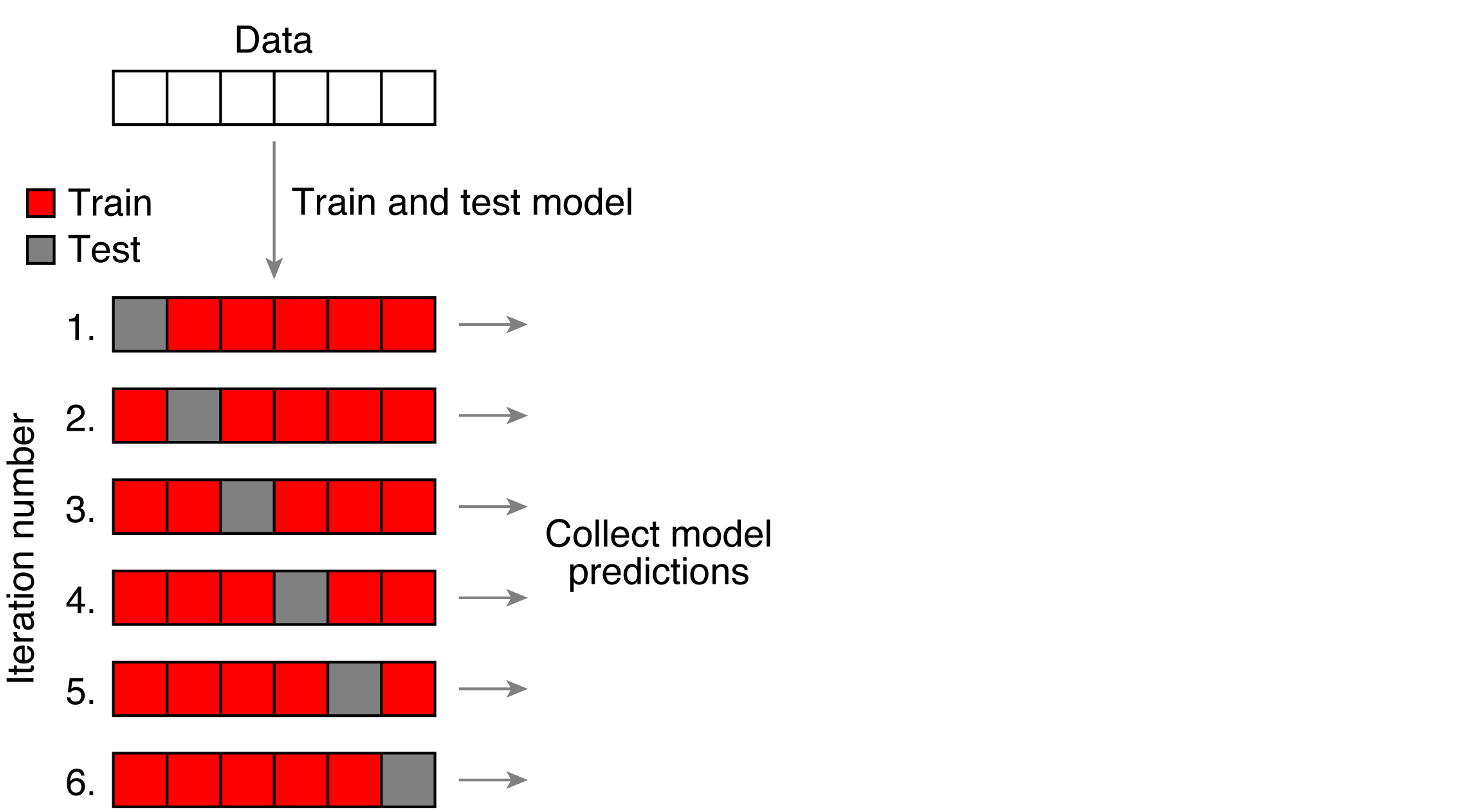
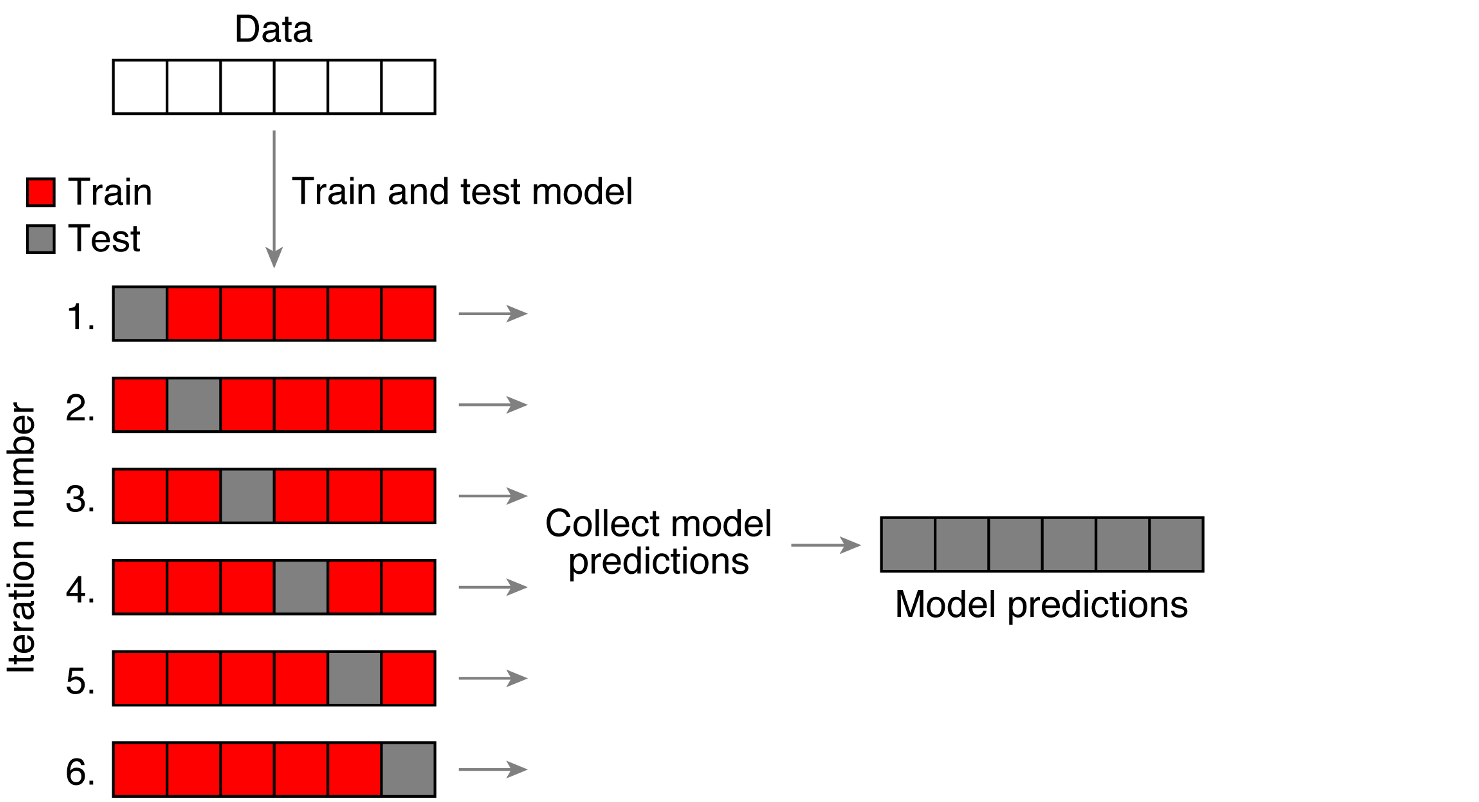
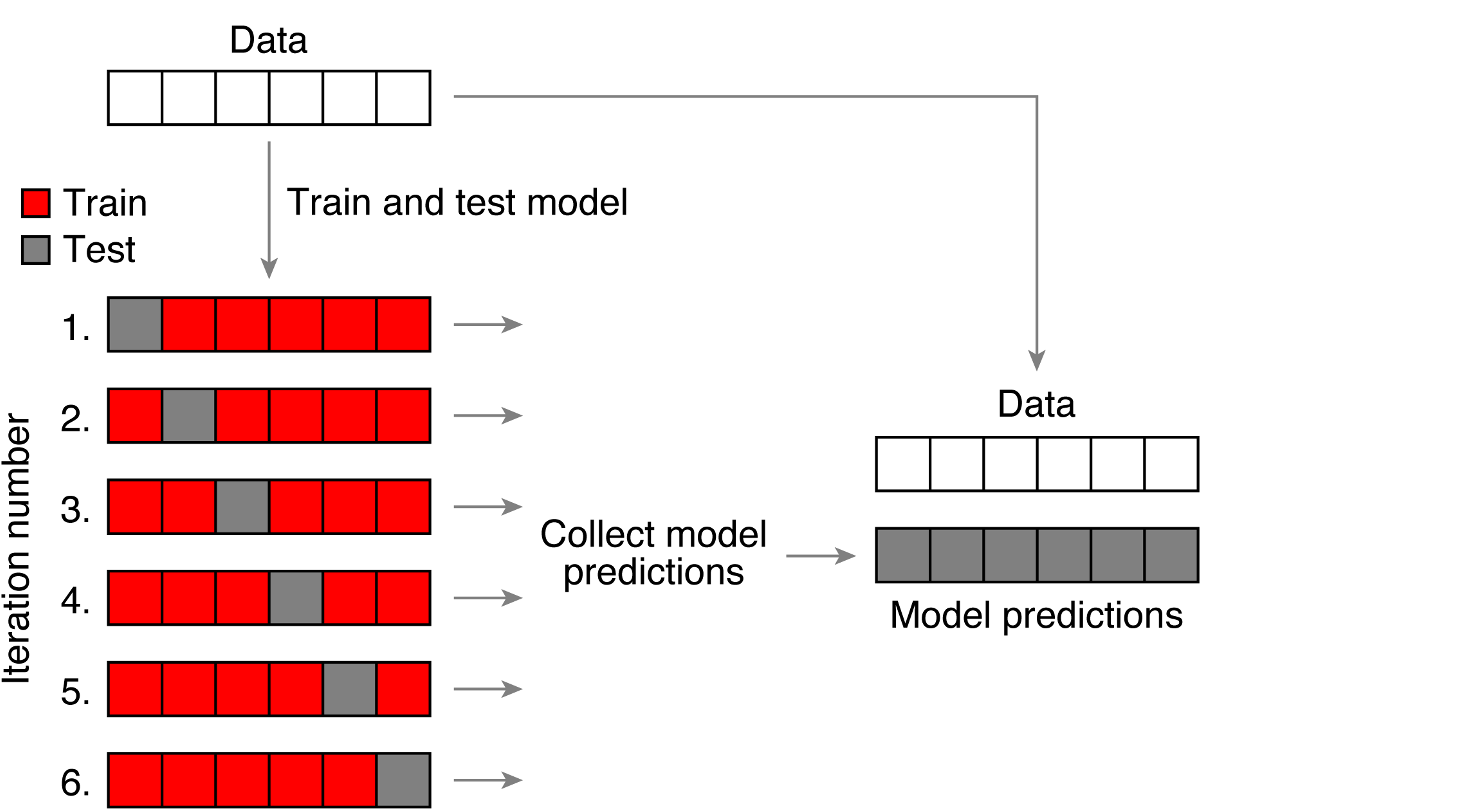
Rows: 4,568
Columns: 36
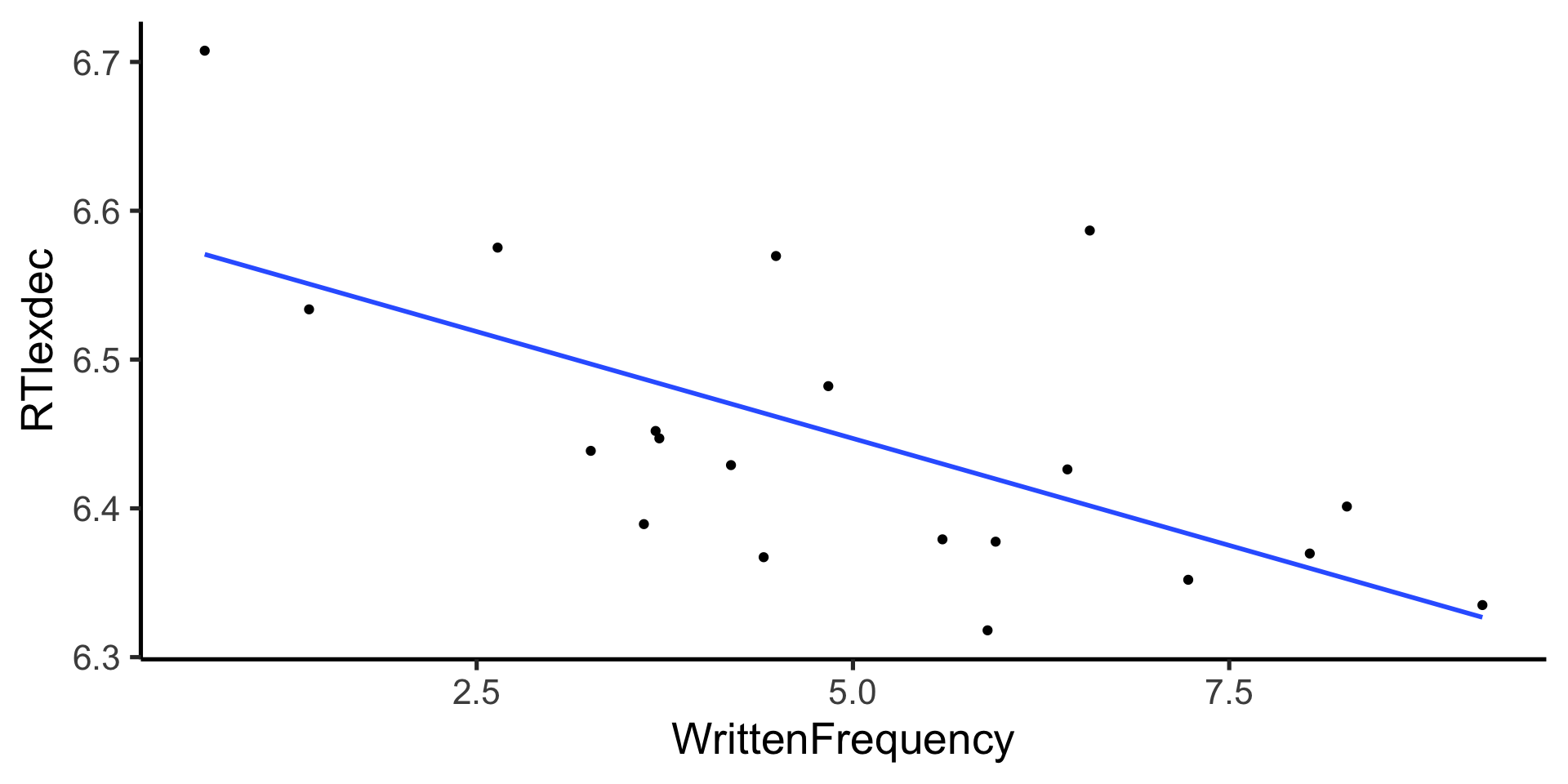
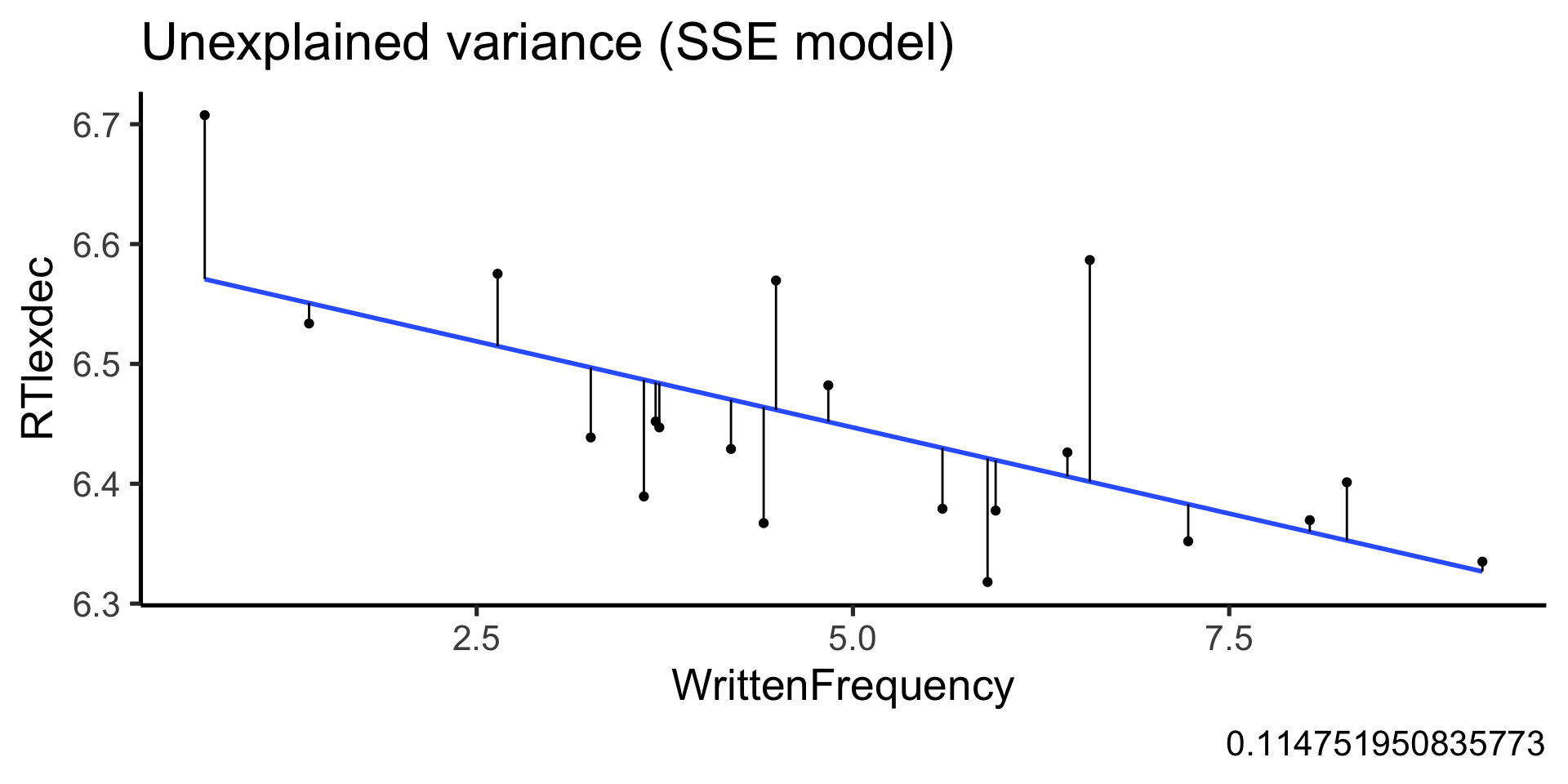
$ RTlexdec <dbl> 6.543754, 6.397596, 6.304942, 6.424221…
$ RTnaming <dbl> 6.145044, 6.246882, 6.143756, 6.131878…
$ Familiarity <dbl> 2.37, 4.43, 5.60, 3.87, 3.93, 3.27, 3.…
$ Word <fct> doe, whore, stress, pork, plug, prop, …
$ AgeSubject <fct> young, young, young, young, young, you…
$ WordCategory <fct> N, N, N, N, N, N, N, N, N, N, N, N, N,…
$ WrittenFrequency <dbl> 3.9120230, 4.5217886, 6.5057841, 5.017…
$ WrittenSpokenFrequencyRatio <dbl> 1.02165125, 0.35048297, 2.08935600, -0…
$ FamilySize <dbl> 1.3862944, 1.3862944, 1.6094379, 1.945…
$ DerivationalEntropy <dbl> 0.14144, 0.42706, 0.06197, 0.43035, 0.…
$ InflectionalEntropy <dbl> 0.02114, 0.94198, 1.44339, 0.00000, 1.…
$ NumberSimplexSynsets <dbl> 0.6931472, 1.0986123, 2.4849066, 1.098…
$ NumberComplexSynsets <dbl> 0.000000, 0.000000, 1.945910, 2.639057…
$ LengthInLetters <int> 3, 5, 6, 4, 4, 4, 4, 3, 3, 5, 5, 3, 5,…
$ Ncount <int> 8, 5, 0, 8, 3, 9, 6, 13, 3, 3, 1, 9, 1…
$ MeanBigramFrequency <dbl> 7.036333, 9.537878, 9.883931, 8.309180…
$ FrequencyInitialDiphone <dbl> 12.02268, 12.59780, 13.30069, 12.07807…
$ ConspelV <int> 10, 20, 10, 5, 17, 19, 10, 13, 1, 7, 1…
$ ConspelN <dbl> 3.737670, 7.870930, 6.693324, 6.677083…
$ ConphonV <int> 41, 38, 13, 6, 17, 21, 13, 7, 11, 14, …
$ ConphonN <dbl> 8.837826, 9.775825, 7.040536, 3.828641…
$ ConfriendsV <int> 8, 20, 10, 4, 17, 19, 10, 6, 0, 7, 14,…
$ ConfriendsN <dbl> 3.295837, 7.870930, 6.693324, 3.526361…
$ ConffV <dbl> 0.6931472, 0.0000000, 0.0000000, 0.693…
$ ConffN <dbl> 2.7080502, 0.0000000, 0.0000000, 6.634…
$ ConfbV <dbl> 3.4965076, 2.9444390, 1.3862944, 1.098…
$ ConfbN <dbl> 8.833900, 9.614738, 5.817111, 2.564949…
$ NounFrequency <int> 49, 142, 565, 150, 170, 125, 582, 2061…
$ VerbFrequency <int> 0, 0, 473, 0, 120, 280, 110, 76, 4, 86…
$ CV <fct> C, C, C, C, C, C, C, C, V, C, C, V, C,…
$ Obstruent <fct> obst, obst, obst, obst, obst, obst, ob…
$ Frication <fct> burst, frication, frication, burst, bu…
$ Voice <fct> voiced, voiceless, voiceless, voiceles…
$ FrequencyInitialDiphoneWord <dbl> 10.129308, 9.054388, 12.422026, 10.048…
$ FrequencyInitialDiphoneSyllable <dbl> 10.409763, 9.148252, 13.127395, 11.003…
$ CorrectLexdec <int> 27, 30, 30, 30, 26, 28, 30, 28, 25, 29…