Data Import and Tidy
Data Science for Studying Language and the Mind
2024-09-10
Announcements
- Course selection period ends today!
- Please email me by 1pm if you require any permits or changes
- Our office admin leaves at 4 and I am not able to see or issue permits myself
- Pset 1 was due yesterday at noon
- Solutions will be posted on Thursday at noon
- If you forgot to turn it in, please email me asap to request an extension to Thursday
- If you are new to the class, please see me after class
- We will try to schedule a remediation day to help you get caught up
Last week
- Data visualization with
ggplot2 - Let’s go back to revisit one thing we missed! Shortcuts
You are here
Data science with R
- R basics
- Data visualization
Data wrangling
Stats & Model buidling
- Sampling distribution
- Hypothesis testing
- Model specification
- Model fitting
- Model accuracy
- Model reliability
More advanced
- Classification
- Inference for regression
- Mixed-effect models
Data science workflow
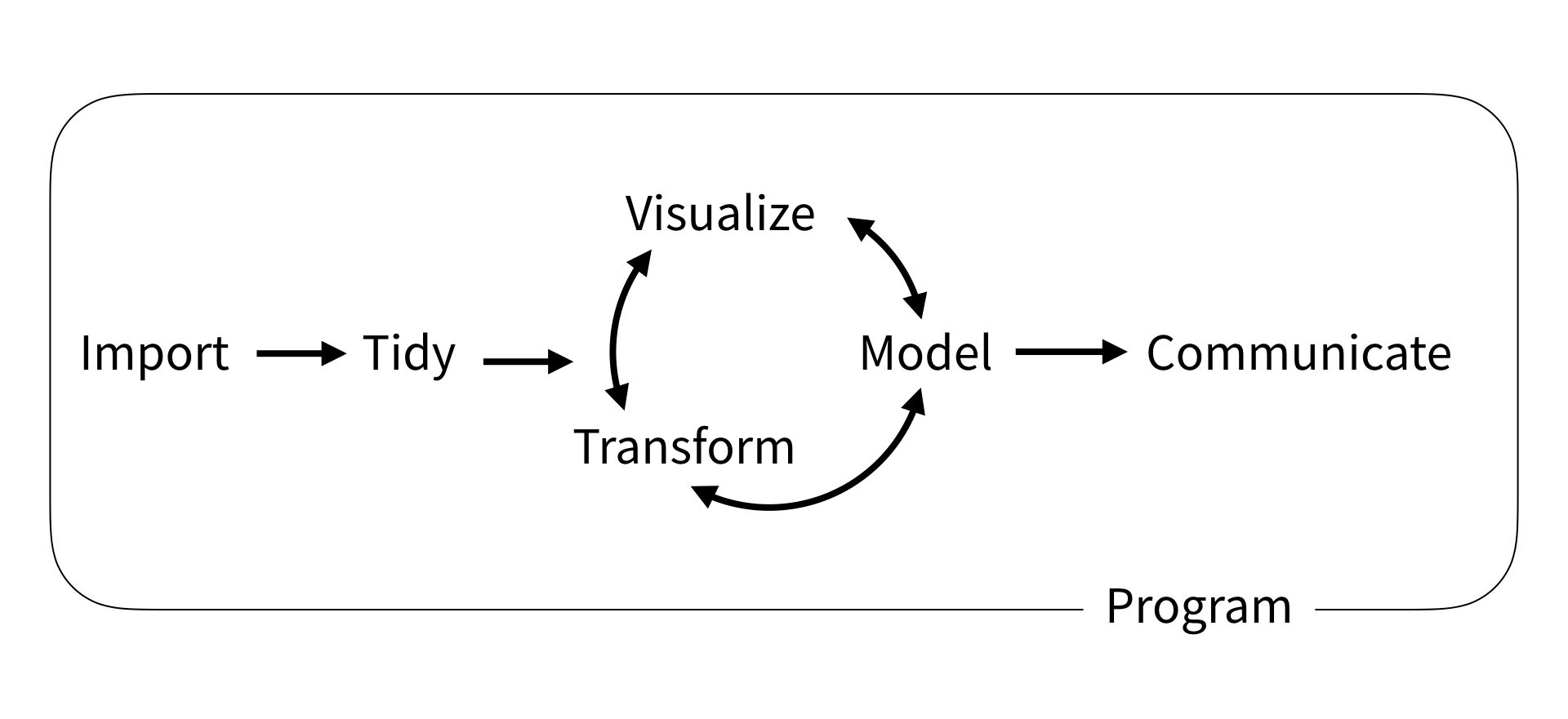
Data Science Workflow by R4DS
Overview for today
- Tidyverse
- Tidy data
purr- functional programmingtibble- modern data.framereadr- reading data
Tidyverse
The tidyverse is an opinionated collection of R packages designed for data science. All packages share an underlying design philosophy, grammar, and data structures.
Tidyverse
ggplot2- for data visualizationdplyr- for data wranglingreadr- for reading datatibble- for modern data framesstringr: for string manipulationforcats: for dealing with factorstidyr: for data tidyingpurrr: for functional programming

Loading the tidyverse
Already installed on Google Colab’s R kernel:
Returns a message in Google Colab:
- a list of packages loaded
- a warning of potential name conflicts
Tidy data
Tidyverse makes use of tidy data, a standard way of structuring datasets:
- each variable forms a column; each column forms a variable
- each observation forms a row; each row forms an observation
- value is a cell; each cell is a single value
Tidy data

Visual of tidy data rules, from R for Data Science
Why tidy data?
- Because consistency and uniformity are very helpful when programming
- Variables as columns works well for vectorized languages (R!)
purr
Functional programming
to illustrate the joy of tidyverse and tidy data
purr
purrr enhances R’s functional programming (FP) toolkit by providing a complete and consistent set of tools for working with functions and vectors. If you’ve never heard of FP before, the best place to start is the family of map() functions which allow you to replace many for loops with code that is both more succinct and easier to read.
The map_*() functions
- Take a vector as input
- Apply a function to each element
- Return a new vector
The map_*() functions
We say “functions” because there are 5, one for each type of vector:
map()- listmap_lgl()- logicalmap_int()- integermap_dbl()- doublemap_chr()- character
map use case
tibble
modern data frames
tibble
A tibble, or tbl_df, is a modern reimagining of the data.frame, keeping what time has proven to be effective, and throwing out what is not. Tibbles are data.frames that are lazy and surly: they do less and complain more
tibble
Tibbles do less than data frames, in a good way:
- never changes type of input (never converts strings to factors!)
- never changes the name of variables
- only recycles vectors of length 1
- never creates row names
Create a tibble
Coerce an existing object:
# A tibble: 4 × 2
x y
<int> <chr>
1 1 a
2 2 b
3 3 c
4 4 d Pass a column of vectors:
Test if tibble
With is_tibble(x) and is.data.frame(x)
data.frame v tibble
You will encounter 2 main differences:
- printing
- by default, tibbles print the first 10 rows and all columns that fit on screen, making it easier to work with large datasets.
- also report the type of each column (e.g.
<dbl>,<chr>)
- subsetting - tibbles are more strict than data frames, which fixes two quirks we encountered when subsetting with
[[and$:- tibbles never do partial matching
- they always generate a warning if the column you are trying to extract does not exist.
readr
reading data
readr
The goal of readr is to provide a fast and friendly way to read rectangular data from delimited files, such as comma-separated values (CSV) and tab-separated values (TSV). It is designed to parse many types of data found in the wild, while providing an informative problem report when parsing leads to unexpected results.
Rectangular data
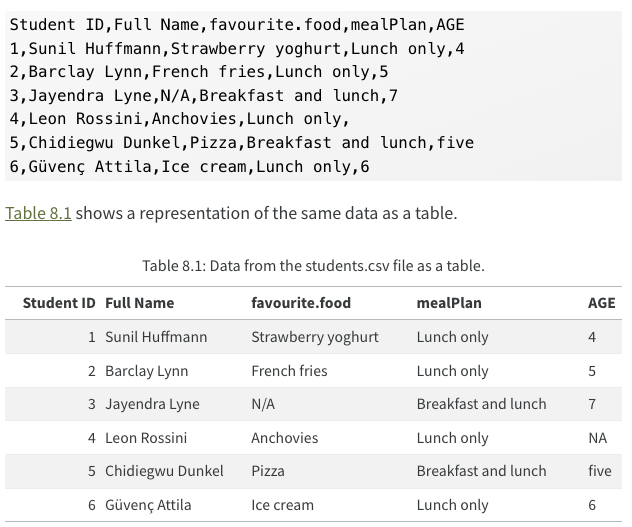
Figure 1: Sample csv file from R for Data Science
Getting a CSV file from google sheets
- Let’s quickly demo how we’d do this from google sheets
read_*()
The read_*() functions have two important arguments:
file- the path to the filecol_types- a list of how each column should be converted to a specific data type
7 supported file types, read_*()
read_csv(): comma-separated values (CSV)read_tsv(): tab-separated values (TSV)read_csv2(): semicolon-separated valuesread_delim(): delimited files (CSV and TSV are important special cases)read_fwf(): fixed-width filesread_table(): whitespace-separated filesread_log(): web log files
Read csv files
Path only, readr guesses types:
col_types column specification
There are 11 column types that can be specified:
col_logical()- reads as boolean TRUE FALSE valuescol_integer()- reads as integercol_double()- reads as doublecol_number()- numeric parser that can ignore non-numberscol_character()- reads as stringscol_factor(levels, ordered = FALSE)- creates factorscol_datetime(format = "")- creates date-timescol_date(format = "")- creates datescol_time(format = "")- creates timescol_skip()- skips a columncol_guess()- tries to guess the column
Reading more complex files
Reading more complex file types requires functions outside the tidyverse:
- excel with
readxl- see Spreadsheets in R for Data Science - google sheets with
googlesheets4- see Spreadsheets in R for Data Science - databases with
DBI- see Databases in R for Data Science - json data with
jsonlite- see Hierarchical data in R for Data Science
Writing to a file
Write to a .csv file with
Common problems readr
Data set containing 3 common problems
# A tibble: 6 × 5
`Student ID` `Full Name` favourite.food mealPlan AGE
<dbl> <chr> <chr> <chr> <chr>
1 1 Sunil Huffmann Strawberry yoghurt Lunch only 4
2 2 Barclay Lynn French fries Lunch only 5
3 3 Jayendra Lyne N/A Breakfast and lunch 7
4 4 Leon Rossini Anchovies Lunch only <NA>
5 5 Chidiegwu Dunkel Pizza Breakfast and lunch five
6 6 Güvenç Attila Ice cream Lunch only 6 - Column contains unexpected values (
AGE) - Missing values are not
NA(AGEandfavorite.food) - Column names have spaces (
Student IDandFull Name)
Column contains unexpected values
Your dataset has a column that you expected to be logical or double, but there is a typo somewhere, so R has coerced the column into character.
# A tibble: 6 × 5
`Student ID` `Full Name` favourite.food mealPlan AGE
<dbl> <chr> <chr> <chr> <chr>
1 1 Sunil Huffmann Strawberry yoghurt Lunch only 4
2 2 Barclay Lynn French fries Lunch only 5
3 3 Jayendra Lyne N/A Breakfast and lunch 7
4 4 Leon Rossini Anchovies Lunch only <NA>
5 5 Chidiegwu Dunkel Pizza Breakfast and lunch five
6 6 Güvenç Attila Ice cream Lunch only 6 Solve by specifying the column type col_double() and then using the problems() function to see where R failed.
Missing values are not NA
Your dataset has missing values, but they were not coded as NA as R expects.
# A tibble: 6 × 5
`Student ID` `Full Name` favourite.food mealPlan AGE
<dbl> <chr> <chr> <chr> <chr>
1 1 Sunil Huffmann Strawberry yoghurt Lunch only 4
2 2 Barclay Lynn French fries Lunch only 5
3 3 Jayendra Lyne N/A Breakfast and lunch 7
4 4 Leon Rossini Anchovies Lunch only <NA>
5 5 Chidiegwu Dunkel Pizza Breakfast and lunch five
6 6 Güvenç Attila Ice cream Lunch only 6 Solve by adding an na argument (e.g. na=c("N/A"))
# A tibble: 6 × 5
`Student ID` `Full Name` favourite.food mealPlan AGE
<dbl> <chr> <chr> <chr> <chr>
1 1 Sunil Huffmann Strawberry yoghurt Lunch only "4"
2 2 Barclay Lynn French fries Lunch only "5"
3 3 Jayendra Lyne <NA> Breakfast and lunch "7"
4 4 Leon Rossini Anchovies Lunch only ""
5 5 Chidiegwu Dunkel Pizza Breakfast and lunch "five"
6 6 Güvenç Attila Ice cream Lunch only "6" Column names have spaces
Your dataset has column names that include spaces, breaking R’s naming rules. In these cases, R adds backticks (e.g. `brain size`);
We can use the rename() function to fix them.
# A tibble: 6 × 5
student_id full_name favourite.food mealPlan AGE
<dbl> <chr> <chr> <chr> <chr>
1 1 Sunil Huffmann Strawberry yoghurt Lunch only 4
2 2 Barclay Lynn French fries Lunch only 5
3 3 Jayendra Lyne N/A Breakfast and lunch 7
4 4 Leon Rossini Anchovies Lunch only <NA>
5 5 Chidiegwu Dunkel Pizza Breakfast and lunch five
6 6 Güvenç Attila Ice cream Lunch only 6 . . . d If we have a lot to rename and that gets annoying, see janitor::clean_names().