Lab 9: Model accuracy
Not graded, just practice
1 Model accuracy
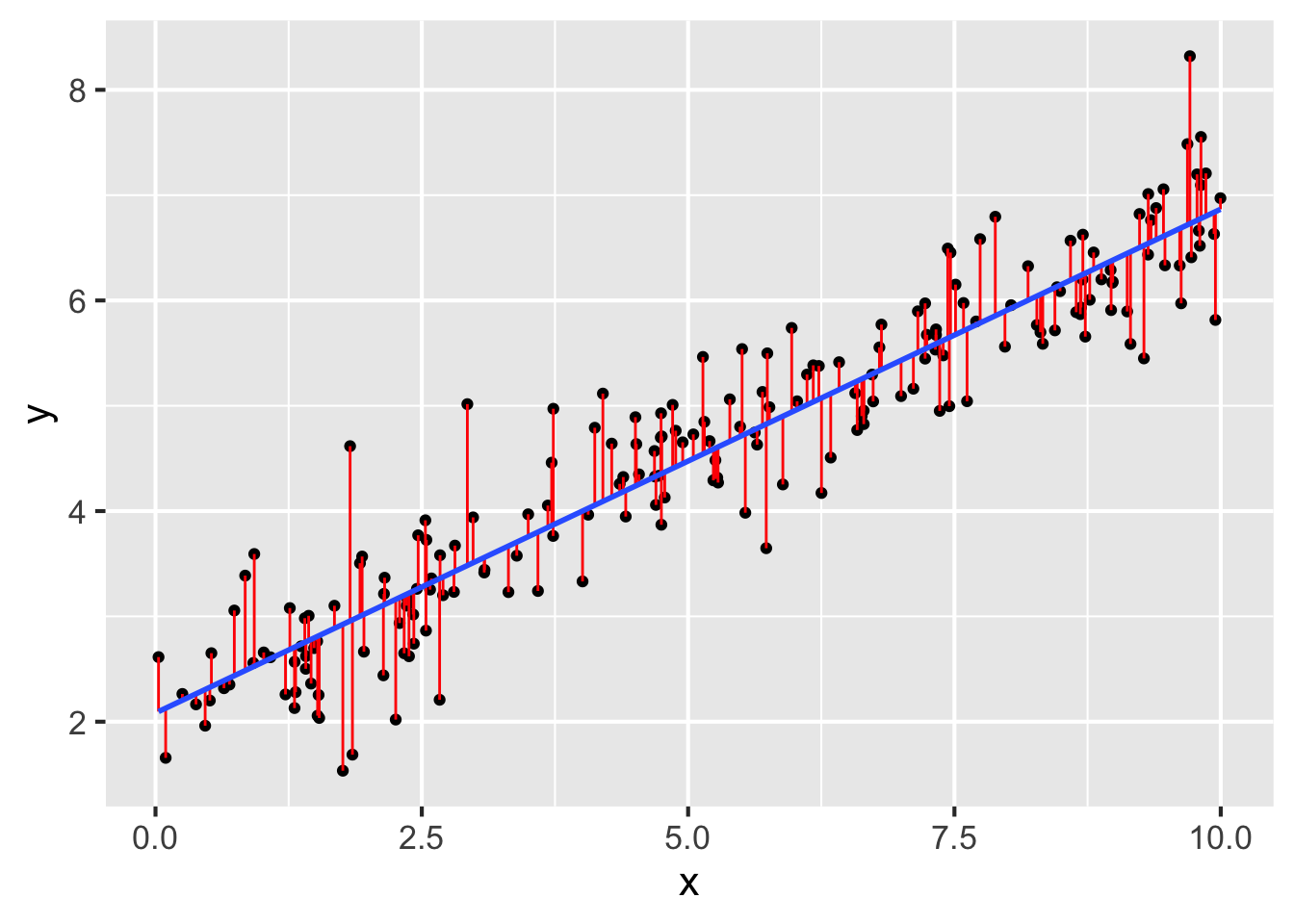
Question 13 refers to the following figure:
- In the figure above, which of the following corresponds to the residuals?
- Suppose the \(R^2\) value on the model in the figure above is about 0.88. Given this value, which of the following best describes the accuracy of the model?
- Suppose 0.88 reflects the \(R^2\) for our fitted model on our sample. Which of the following is true about the \(R^2\) for our fitted model on the population?
- Which of the following best describes an overfit model?
- How can we estimate \(R^2\) on the population? Choose all that apply.
- Fill in the blanks below to best describe cross validation:
- Leave some out
- Fit a model on the data
- Evaluate the mdoel on the data
2 Model accuracy in R
Questions 19-20 refer to the following code and output:
model <- lm(y ~ 1 + x, data)
summary(model)
Call:
lm(formula = y ~ 1 + x, data = data)
Residuals:
Min 1Q Median 3Q Max
-1.38959 -0.32626 -0.04605 0.31967 1.65977
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.08280 0.07418 28.08 <2e-16 ***
x 0.47844 0.01242 38.51 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.5139 on 198 degrees of freedom
Multiple R-squared: 0.8822, Adjusted R-squared: 0.8816
F-statistic: 1483 on 1 and 198 DF, p-value: < 2.2e-16- What is the \(R^2\) value for the model fit above?
- Does the value you entered in 19 reflect \(R^2\) on the population or on the sample?
Questions 21-23 refer to the following code and output:
# we divide the data into v folds for cross-validation
set.seed(2)
splits <- vfold_cv(data)
# model secification
model_spec <-
linear_reg() %>%
set_engine(engine = "lm")
# add a workflow
our_workflow <-
workflow() %>%
add_model(model_spec) %>%
add_formula(y ~ x)
# fit models to our folds
fitted_models <-
fit_resamples(
object = our_workflow,
resamples = splits
)
fitted_models %>%
collect_metrics()# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 rmse standard 0.507 10 0.0397 Preprocessor1_Model1
2 rsq standard 0.890 10 0.0146 Preprocessor1_Model1- In the cross-validation performed above, how many folds were the data split into?
- What \(R^2\) do we estimate for the population?
- What model has been fit?
Questions 24-26 refer to the following code and output:
# we bootstrap the data for cross-validation
set.seed(2)
bootstrap <- bootstraps(data)
# fit models to our folds
fitted_models_boot <-
fit_resamples(
object = our_workflow,
resamples = bootstrap
)
fitted_models_boot %>%
collect_metrics()# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 rmse standard 0.507 25 0.00946 Preprocessor1_Model1
2 rsq standard 0.887 25 0.00377 Preprocessor1_Model1- How many bootstrap samples did we generate?
- True or false, we fit the same model to the bootstrap data as we did in the cross-validation code.
- True or false, the \(R^2\) estimated by bootstrapping is equal to the \(R^2\) estimated by cross-validation.